Pool testing on random and natural clusters of individuals: optimisation of SARS-CoV-2 surveillance in the presence of low viral load samples
Publication date: 23/10/2020 – E&P Code: repo.epiprev.it/1990
Authors: Michela Baccini1,2, Alessandra Mattei1,2, Irene Paganini3, Emilia Rocco1,2, Cristina Sani3, Giulia Vannucci1,2, Simonetta Bisanzi3, Elena Burroni3, Marco Peluso3, Armelle Munnia3, Filippo Cellai3, Giampaolo Pompeo3, Laura Micio3, Jessica Viti3, Fabrizia Mealli1,2, Francesca Carozzi3
Abstract: Facing the SARS-CoV-2 epidemic requires intensive testing on the population to early identify and isolate infected subjects. During the emergency phase of the epidemic, RT-qPCR on nasopharyngeal (NP) swabs, which is the most reliable technique to detect ongoing infections, exhibited limitations due to availability of reagents and budget constraints. This stressed the need to develop screening procedures requiring fewer resources and suitable to be extended to larger proportions of the population. RT-qPCR on pooled samples from individual NP swabs seems to be a promising technique to improve surveillance.
We performed preliminary experimental analyses aimed to investigate the performance of pool testing on samples with low viral load and we evaluated through Monte Carlo (MC) simulations alternative screening protocols based on sample pooling, tailored to contexts characterized by different infection prevalence. We focused on the role of pool size and the opportunity to take advantage of natural clustering structure in the population, e.g. families, school classes, hospital rooms.
Despite the use of a limited number of specimens, our results suggest that, while high viral load samples seem to be detectable even in a pool with 29 negative samples, positive specimens with low viral load may be masked by the negative samples, unless smaller pools are used. The results of MC simulations confirm that pool testing is useful in contexts where the infection prevalence is low. The gain of pool testing in saving resources can be very high, and can be optimized by selecting appropriate group sizes. Exploiting natural groups makes it convenient the definition of larger pools and potentially overcomes the issue of low viral load samples by increasing the probability of identifying more than one positive in the same pool.
Cite as: Michela Baccini, Alessandra Mattei, Irene Paganini, et. al. (2020). Pool testing on random and natural clusters of individuals: optimisation of SARS-CoV-2 surveillance in the presence of low viral load samples. E&P Repository https://repo.epiprev.it/1990
Topic: COVID-19
Key words: cluster naturali, costi-efficacia, COVID-19, RT-qPCR test, SARS-CoV-2, screening,
AVVERTENZA. GLI ARTICOLI PRESENTI NEL REPOSITORY NON SONO SOTTOPOSTI A PEER REVIEW.
Info
Affiliations:
1 Department of Statistics, Computer Science, Applications, University of Florence, Florence, Italy
2 Florence Center for Data Science, University of Florence, Florence, Italy
3 Regional Laboratory of Cancer Prevention, Institute for prevention, research and oncological network (ISPRO), Florence, Italy
Authors’ contributions: –
Competing interests: none.
Funding disclosure: none.
Ethics committee approval: -.
Copyright: Il detentore del copyright è l’autore/finanziatore, che ha concesso a “E&P Repository” una licenza per rendere pubblico questo preprint. The copyright holder for this preprint is the author/funder, who has granted E&P Repository a license to display the preprint in perpetuity.
Terms of distribution: CC BY-NC-ND
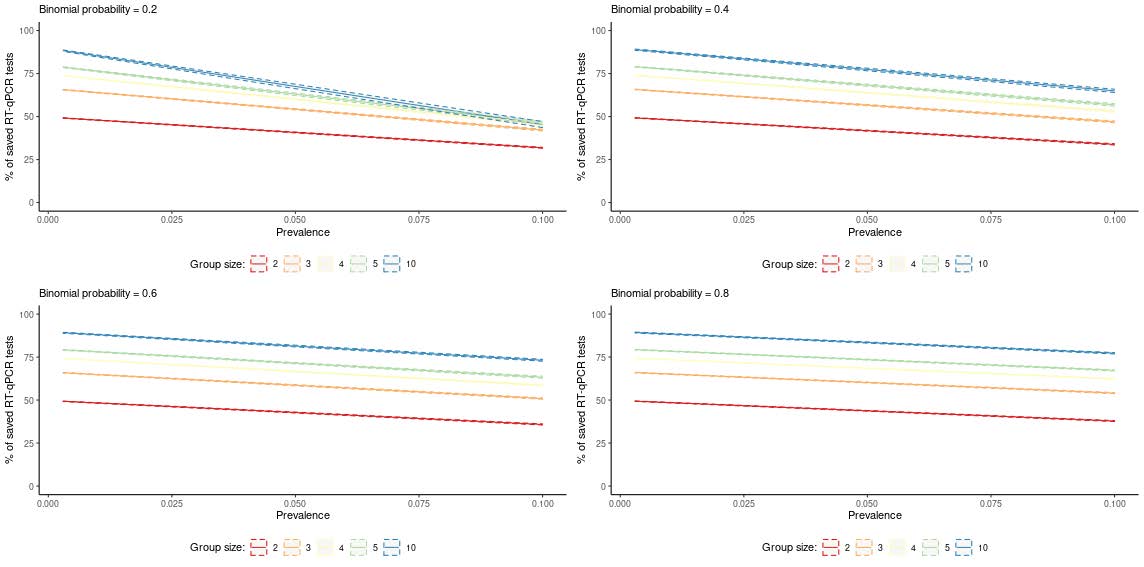
Figure 2. Pool testing on natural groups: Monte Carlo means (solid lines) and 90% variability intervals (dashed lines) of the percentage of saved RT-qPCR tests by prevalence of infection in the population (p), group size (k) and Binomial probability (π) . Population size = 10,000. Specificity of pool testing = 0.997; Sensitivity of pool testing = 0.995
References & Citations
Google Scholar

