A SIRD model calibrated on deaths to investigate the second wave of the SARS-CoV-2 epidemic in Italy
Publication date: 05/12/2020 – E&P Code: repo.epiprev.it/2052
Authors: Giulia Cereda1, Cecilia Viscardi1, Luca Gherardini1, Fabrizia Mealli1, Michela Baccini1
Abstract: After the SARS-CoV-2 outbreak during the 2020 spring, Italy faced a second epidemic wave during the autumn. Using a SIRD model calibrated on the COVID19-related deaths, we describe the epidemic dynamics from August 1st to November 30th 2020, region by region. We explore the behavior of the contagion in terms of time-varying reproductive number R0 and estimated number of circulating infections. This number, when compared to the number of notified positives, provides an evaluation of the submerged portion of contagion. The results indicate that in Italy, during the second SARS-CoV-2 epidemic wave, the reproductive number changed over time heterogeneously across regions, but with some important common elements including a mid-October peak and a decline during the month of November, which are visible in most regions. Ad hoc studies should be performed to investigate the causal effect that specific events (e.g. schools reopening, regional elections) and restrictions of different degree have had on inflating or deflating the rate of contagion. Despite the decline of R0(t) in most regions, the prevalence of circulating infections estimated at the end of the study period is not negligible, in particular in the North of the country. This suggests that even small future increases of R0(t) may lead in a short time to levels of contagion spread that could be unsustainable, depending on the regional supply of hospital and intensive care unit beds and the organizations of healthcare services throughout the territory.
Cite as: Giulia Cereda, Cecilia Viscardi, Luca Gherardini, Fabrizia Mealli, Michela Baccini (2020). A SIRD model calibrated on deaths to investigate the second wave of the SARS-CoV-2 epidemic in Italy. E&P Repository https://repo.epiprev.it/2052
Topic: COVID-19
Key words: COVID-19, infection fatality rate, parametric boostrap, regioni italiane, reproductive number, SARS-CoV-2, seconda ondata epidemica, SIRD,
AVVERTENZA. GLI ARTICOLI PRESENTI NEL REPOSITORY NON SONO SOTTOPOSTI A PEER REVIEW.
Info
Affiliations:
1 Department of Statistics, Computer Science, Applications, University of Florence, Florence Center for Data Science, University of Florence
Authors’ contributions:
GC conceptualized the problem, wrote the code, performed the statistical analyses, contributed in discussing the results, contributed in writing the paper.
CV conceptualized the problem, contributed in writing the code, contributed in writing the paper.
LG contributed in writing the code, contributed in performing the statistical analyses.
FM contributed in discussing the results, contributed in writing the paper.
MB conceptualized the problem, contributed in writing the code, discussed the results, wrote the first version of the paper, supervised.
All authors read and approved the final version of the manuscript.
Competing interests: none
Funding disclosure: none
Ethics committee approval: The study was based on publicly available aggregate data. No Ethics committee approval was necessary.
Copyright: Il detentore del copyright è l’autore/finanziatore, che ha concesso a “E&P Repository” una licenza per rendere pubblico questo preprint. The copyright holder for this preprint is the author/funder, who has granted E&P Repository a license to display the preprint in perpetuity.
Terms of distribution: CC BY-NC-ND
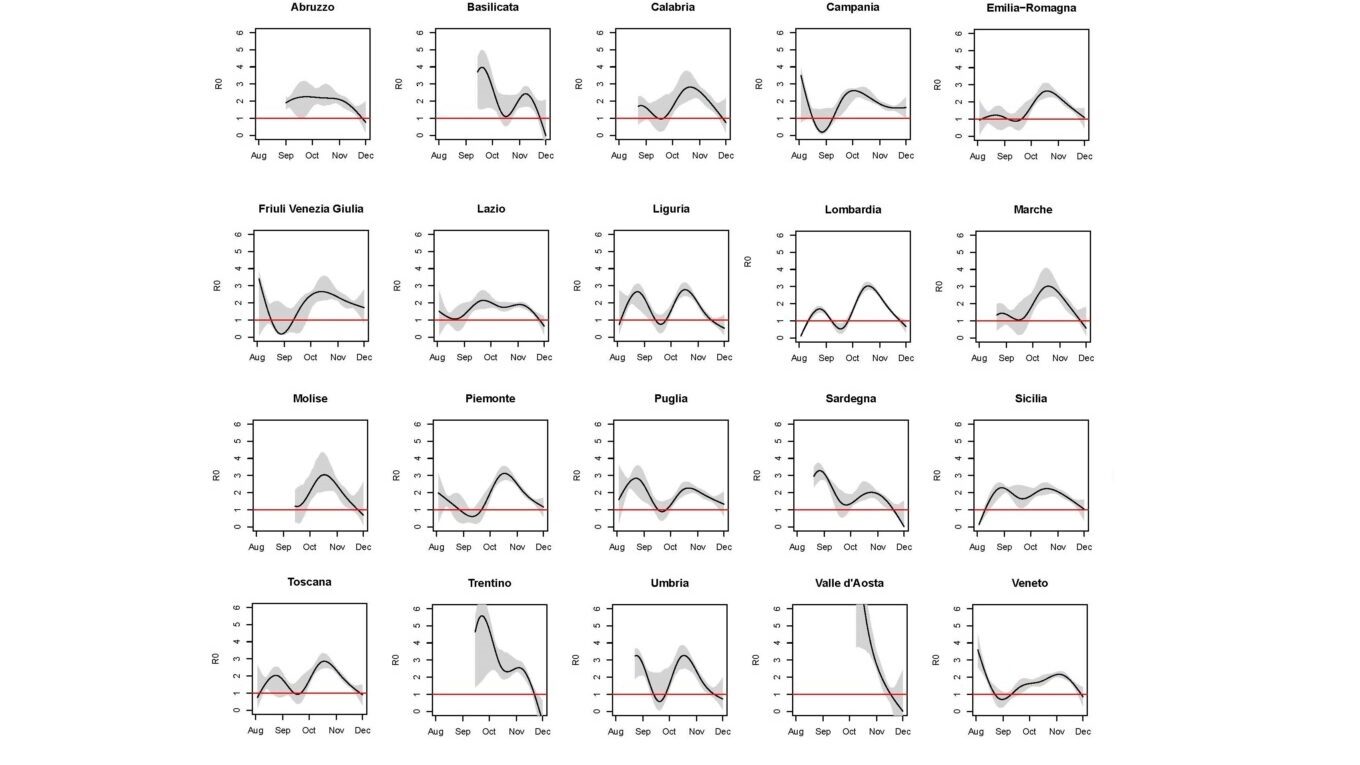
Estimated R₀(t) with pointwise 90% confidence bands assuming an infection fatality rate equal to 0.5%, by region
References & Citations
Google Scholar

